Researchers around the world have access to a greater variety and volume of genomics data than ever before. Genomics is now available to a vast majority of researchers, pushing forward the discovery at a tremendous pace and changing people’s lives. This growth is happening because of the perfect storm between genomic testing and technological improvements. In the span of a few decades, the cost of human genome sequencing has gone from millions of dollars to hundreds of dollars.
At Microsoft, we recognize the challenges faced by the genomics community and are striving to build an ecosystem (backed by open source and Microsoft products and services) that can facilitate genomics computing work for all. We’ve focused our efforts on 3 main core areas: research and discovery in genomics data, building out a platform to enable rapid automation and analysis at scale, and optimized and secure pipelines at a clinical level. One of the core Azure services that has enabled us to leverage an HPC environment to perform the genomic analysis is Azure CycleCloud.
Genomic analysis at scale requires hyperscale compute
Sequencing a single individual’s genome, or even that of a small cohort of individuals, generates a significant amount of data and requires a massive amount of computational power to analyze. The computing power required to effectively analyze, share, and disseminate this data has historically been constrained by what could be provided on-premises for research organizations. For most researchers, the scarcity of availability of high performance computing (HPC) clusters has hamstrung their research potential, threatening high upfront infrastructure investments and long-term maintenance costs. Furthermore, to capture an accurate representation of population health, research studies today need to be globalized meaning that genomic data needs to be securely stored, shared, and transmitted across the world—creating a computing demand that is a heavy lead for even the most sophisticated on-premises. As such, the lack of adequate computing technology has delayed and constrained the ability of genomic research communities to easily collaborate and share findings.
Cloud computing and the broader digital transformation of the health industry have been powerful enablers of modern genomic breakthroughs, unleashing a practically limitless—and more widely and affordably available—ability to meet the computing demands needed by research organizations and medical institutions to advance genomic science.
HPC on Azure and Azure CycleCloud for genomic analysis
Azure CycleCloud is an enterprise-friendly tool for orchestrating and managing HPC environments on Azure. With Azure CycleCloud, users can provision infrastructure for HPC systems, deploy familiar HPC schedulers, and automatically scale the infrastructure to run jobs efficiently at any scale. Through Azure CycleCloud, users can create different types of file systems and mount them to the compute cluster nodes to support HPC workloads. With dynamic scaling of clusters, the business can get the resources it needs at the right time and the right price, with Azure CycleCloud’s automated configuration enabling IT to ultimately focus on providing high-value services to the business users.
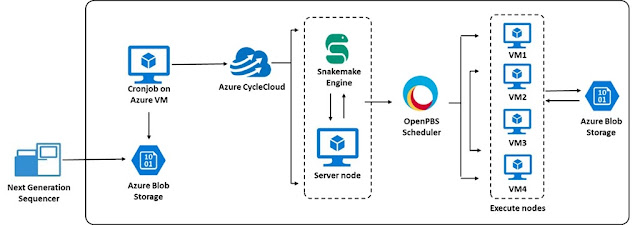
Workflow managers (like Cromwell, Galaxy, Nextflow, and Snakemake) are used for accelerating genome analysis by making them more efficient and scalable. For instance, a typical next-generation sequencing machine can sequence anywhere from 12 to 192 samples per run and creates an output file (called Binary Base Call [BCL] are the raw data generated by NGS). This output file is converted into several FastQ files (FastQ file is a text-based format for storing both a nucleotide sequence and its corresponding quality score). Each FastQ must be further converted into BAM Format (a binary format for storing sequence data) and then Variant Call Format (VCF), which specifies the format of a text file used in bioinformatics for storing gene sequence variations. A bioinformatician or a clinical scientist then picks these files up for further analysis. The sequence of conversion steps from BCL to FASTQ files can take between several hours to several days on commodity hardware. One way to drastically reduce this timeline is to use Azure CycleCloud or Azure Batch to configure these steps as a set of jobs that can be run in parallel.
Accelerating germline testing by optimizing secondary analysis using Azure CycleCloud
Belfast Health and Social Care Trust is the largest integrated health and social care trust in the United Kingdom. They deliver integrated health and social care services to approximately 340,000 citizens in Belfast and provide many regional specialist services to all of Northern Ireland. Belfast Trust also comprises the major network of teaching and training hospitals in Northern Ireland.
Within Belfast Trust, the Regional Molecular Diagnostics Service Northern Ireland (RMDS) has been funded to develop and deliver a service that provides molecular testing of germline and somatic disorders through the introduction of a comprehensive portfolio of Next Generation Sequencing (NGS) panels and exomes aimed at improving patient outcomes in Northern Ireland.
The main goal of this strategic initiative is to enhance high-quality patient care by improving the turnaround time for genomic analysis in accordance with the best practice guidelines by The Association for Clinical Genomic Science and delivering an equitable molecular service in line with mainland United Kingdom labs.
Initially, the sheer complexity and size of the genomic data generated were considered a major computational barrier to the delivery of the service. To meet the computational demand, Belfast Trust developed an accredited germline computational pipeline using the Snakemake workflow manager on Azure. The initial pipeline analyzed only targeted panels, which are smaller in size than clinical exomes, whole exomes, or whole genomes. To convert the raw data to the format necessary for genomic analysis, the jobs were configured for sequential execution on a single virtual machine.






0 comments:
Post a Comment