Genomics is foundational to the development of targeted therapeutics and precision medicine. Advances in DNA sequencing technologies has driven a revolution in genomics-based research and is helping facilitate better understanding of human biology and disease conditions. This expanded knowledge is leading to the proliferation of personalized medicine strategies to prevent, diagnose, and treat diseases. The trend will continue to accelerate in the coming decade as use of genomics information becomes central to clinical decision support and healthcare delivery.
Sequencing genomes at the population level will be required to decipher the genomic fingerprint of a disease, predict interpersonal variability in progression and treatment response, and develop models for clinical decision support. The resulting explosion in genomics data and the computational power required for analysis (tens of exabytes and trillions of core hours in the next five years1) will require agility, easier management, data security, and access to scalable storage and compute capacity.
The demand for cloud-based solutions is evident. It is increasingly being recognized that community driven standards and open-source tools will be necessary in enabling data accessibility, tool interoperability, and reliability of results and models. Microsoft not only supports open-standards and open-source projects but has been actively contributing to these community driven efforts by making it easier to use these tools and software on Azure.
To that end, Microsoft Genomics has released several open source projects on GitHub, including Cromwell on Azure, Genomics Notebooks and Bioconductor support for Azure. We have also made available a growing list of genomics public datasets on the Azure Open Dataset platform.
Scale and automate genomic workflows on Azure with Cromwell
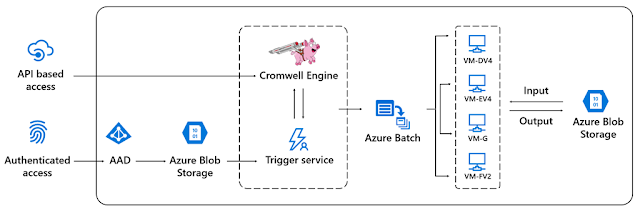
Cromwell is an open-source workflow management system geared toward scientific workflows, originally developed by the Broad Institute. With Cromwell on Azure, users can accelerate their genomic research with the hyperscale compute capabilities of Azure. Cromwell orchestrates the dynamic provisioning of computing resources via Azure Batch and integrates with customers’ Azure Blob storage account for easy data access.
Propelling novel next generation sequencing (NGS) based detection and characterization assay for COVID-19 with Biotia
Biotia is an emerging startup focused on building a platform leveraging next-generation DNA sequencing (NGS) and artificial intelligence (AI) for precision disease detection and diagnosis. They were looking for a cloud-based workflow solution to manage their NGS pipelines and Cromwell on Azure was able to meet their key requirements.
"At Biotia, we have achieved substantial parallelization, thorough version control, and novel COVID-19 detection results by using Cromwell on Azure to back our compute-intensive genomics workflows. We are pleased to include Cromwell on Azure in our bioinformatics software stack." —Joe Barrows, Director of Software Engineering at Biotia
Enable collaborative and repeatable data analysis using Genomics Notebooks powered by Jupyter Notebooks on Azure
Jupyter Notebooks provides users an environment for analyzing data using R or Python and enabling reusability of methods and reproducibility of results. Biomedical researchers and data scientists are increasingly using notebooks for their genomics data analysis needs and for building machine learning models based on multi-modal datasets (genomic, phenotypic, clinical, EMR, demographic, etc.).
Microsoft’s Genomics Notebooks open-source project provides a growing collection of pre-configured notebooks that users can easily launch and use in their Azure workspace. These pre-configured notebooks cover scenarios from genomics variant detection, filtration, annotation, to transformation of the genomic, phenotypic and clinical data into multi-modal data frames needed for data querying and building machine learning models.
Leveraging genomic data to assess the impact of environmental change with the Canadian Department of Fisheries and Oceans
The Canadian Department of Fisheries and Oceans (DFO) is responsible for preserving Canada’s aquatic natural resources. DFO researchers at the Bedford Institute of Oceanography in Dartmouth, Nova Scotia have been using genomics to understand the impact of climate change and human activity on the migration patterns, genetic diversity and population demography of fish such as Atlantic Salmon and Atlantic Cod, which can have major socio-economic implications for the communities that rely on these resources.
The research teams are starting to sequence fish genomes in the hundreds and were looking for Azure based solutions for scaling and streamlining their growing genomics and data analysis needs. The team successfully deployed, and scale tested Cromwell on Azure and is now looking to adopt it as a common genomics workflow platform across their various institutions.
“Leveraging Cromwell on Azure for running our genomics pipelines give us the ability to scale our analysis to thousands of genomes for any species of fish with automation. We can essentially eliminate three months’ time of manual work to generate all the variant calls we need and move directly into connecting that data with other data sources we have. The data science tools will help us easily build and train complex multi-modal data models to gain deeper insights into the impact resulting from interactions between genetic factors, climate information, and human impacts on these species, and predict how they might respond to environmental challenges in the future.” —Dr. Tony Kess, Researcher at the Bradbury Population Genomics Lab, a part of the Bedford Institute of Oceanography in Dartmouth, Nova Scotia
Easily access the vast collection of community-built bioinformatics tools with Bioconductor on Azure
Bioconductor is an open source, open development project that focuses on providing a repository of extensible statistical and graphical software packages, developed in R, for the analysis of high-throughput genomic and biomedical data. Microsoft is collaborating with the Bioconductor core team in bringing Azure support for this wide-ranging OSS software repository.
Bioinformaticians and data scientists can now easily use their preferred Bioconductor software packages on Azure by deploying the preconfigured Bioconductor Docker image hosted in the Microsoft Container Registry on Docker Hub. Additionally, users can also use Azure Virtual Machine (VM) templates to deploy Genomics Data Science VM preconfigured with popular tools for data exploration, analysis, machine learning, and deep learning model development.
Power data analysis and machine learning models with genomics datasets available through the Azure Open Data platform
The Genomics Data Lake on the Azure Open Dataset platform provides a growing compendium of curated and publicly available genomics datasets. These datasets have been generated by key international collaborative efforts with a focus on providing resources for the biomedical research community. Users across healthcare, pharma and life sciences can now use the Genomics Data Lake on Azure to access these datasets for free and easily integrate the data into their genomics analysis workflows.
Accelerate whole exome and genome processing using Microsoft Genomics turnkey service on Azure
Microsoft Genomics is a highly scalable Azure service to perform secondary analysis of the human genome using the Burrows-Wheeler Aligner (BWA) and the Genome Analysis Toolkit (GATK) open-source software. The service is ISO-certified, enables customers’ compliance with HIPAA, and is covered under the Microsoft Business Associate Agreement (BAA). Microsoft continues to optimize the performance of the service by leveraging the innovations in Azure’s high-performance compute infrastructure enabling customers to generate durable genetic variant data from whole genome sequence data (WGS) within hours. Compliance, performance, data durability and provenance make the service ideal for integration into genomics-based clinical decision support workflows.
Accelerating scientific discoveries to advance cures for childhood cancers through access to real-time clinical genome sequencing at St. Jude Children’s Research Hospital
Whole-genome sequencing offers the most comprehensive assessment of differences between patients’ normal and cancer genomes. Realtime access to the genomic information is not only important for clinical decision support, but it can also accelerate research and novel discoveries and cures. St. Jude Children’s Research Hospital has partnered with Microsoft and DNAnexus to build St. Jude Cloud—the world’s largest public repository of pediatric genomics data.
This first-of-its-kind initiative provides researchers from around the world access to high-quality whole-genome, whole-exome and transcriptome data from appropriately consented St. Jude patients who have undergone clinical genomic profiling. St. Jude Cloud uses Azure and the Microsoft Genomics service to quickly upload, analyze, and harmonize the genomics data, which is subsequently made available through the St. Jude Cloud data browser to researchers worldwide.
“Access to high-quality clinical genomic data, generated leveraging the Microsoft Genomics service and streamed to St. Jude Cloud, will help further research in precision medicine for childhood cancer and other diseases.”—Dr. Jinghui Zhang, Chair of Department of Computational Biology at the St. Jude Children’s Research Hospital
Source: azure.microsoft.com






0 comments:
Post a Comment